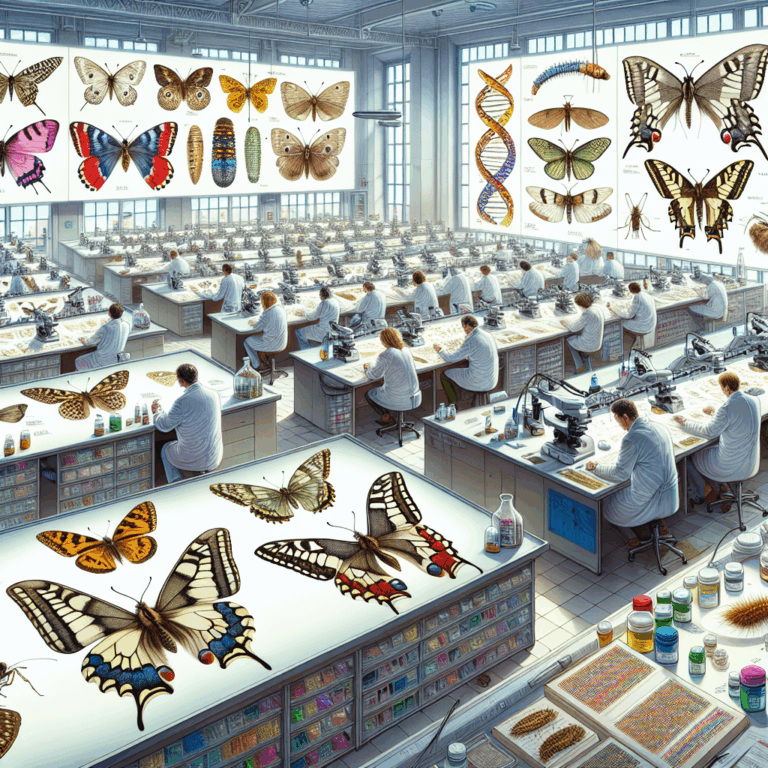
It’s a little after 6:30 on a brisk July morning in a stone hut high in the Italian Alps. A gently hissing wood fire is leaking some warmth out of a brick oven. Gathered near it, around a big wooden table, some of Europe’s brightest young lepidopterists are doing what they do best: arguing in Spanish, Italian, and English about moths.
 The Alte Pforzheimer Hütte, a stone house originally built in 1901, served as a base camp for the lepidopterists hunting rare moths in the Italian Alps.Luigi Avantaggiato
The Alte Pforzheimer Hütte, a stone house originally built in 1901, served as a base camp for the lepidopterists hunting rare moths in the Italian Alps.Luigi Avantaggiato
Scattered across the top of the table are dozens of moths in plastic specimen jars, the harvest of the previous night’s trapping. At one end of the table, Gioele Moro of the Czech Academy of Sciences is gently prying loose moths from the depths of a trap. At the other end, Laura Torrado-Blanco of the University of Oviedo’s entomological collection is paging through Lepidoptera guide books. She’s using the books to identify species—up here at 2,300 meters, there is no Internet connection.
 A few of the scores of moths captured on a single night at a site in the Italian Alps are lined up on a bench in the stone hut. Researchers will identify the moths’ species and some of the insects will be sent on for tissue sampling and eventual genome sequencing. Luigi Avantaggiato
A few of the scores of moths captured on a single night at a site in the Italian Alps are lined up on a bench in the stone hut. Researchers will identify the moths’ species and some of the insects will be sent on for tissue sampling and eventual genome sequencing. Luigi Avantaggiato
Looking up from a book, she notices me noticing the big butterfly tattoo on her left arm. “Chapman’s ringlet,” she tells me. “Erebia palarica,” she adds reflexively.
Pep Lancho Silva, a doctoral student at the Institute of Evolutionary Biology in Barcelona, extends a finger toward me with a spectacular creature on it: a large bone-white moth, with a black head and big black splotches on its wings. Torrado-Blanco is pretty sure it’s Arctia flavia, a species of tiger moth found only in rarefied air. If so, it’s precisely the kind of insect they came up here, to this chilly hut on the edge of a crystalline Alpine pond, to capture.
 A yellow tiger moth, Arctia flavia, is among the catch at the stone hut, at an altitude of 2,300 meters.
A yellow tiger moth, Arctia flavia, is among the catch at the stone hut, at an altitude of 2,300 meters.
 At the crack of dawn in the stone hut, researchers [from left] Eric Toro Delgado, Laura Torrado-Blanco, Mónica Doblas-Bajo, and Gioele Moro (standing) unpack and examine the moths captured during the previous night.Luigi Avantaggiato
At the crack of dawn in the stone hut, researchers [from left] Eric Toro Delgado, Laura Torrado-Blanco, Mónica Doblas-Bajo, and Gioele Moro (standing) unpack and examine the moths captured during the previous night.Luigi Avantaggiato
Lepidopterists have trapped, identified, and classified moths and butterflies for centuries. But this high-altitude confab is no Victorian perambulation. It’s a vital component of a sprawling, cutting-edge project that is pushing the boundaries of bioinformatics and the tools of modern genomics. These researchers are taking part in the first international field expedition of Project Psyche, whose goal is to sequence the genomes of all 11,000 species of moths and butterflies in Europe. Psyche is part of a larger effort, the Darwin Tree of Life project, which is itself a component of arguably the most ambitious science project of all time: the Earth BioGenome Project. Its goal is to sequence the genomes of all of Earth’s roughly 1.8 million organisms—every named species of animal, plant, fungus, and microbe that’s made up of cells that have a nucleus.
Related: The Quest to Sequence the Genomes of Everything
None of these hugely ambitious efforts would be conceivable without the enormous advances in genome sequencing and bioinformatics over the past couple of decades. The cost and speed of sequencing an individual genome have declined to the point where it’s now possible to batch process multiple genomes in a single day and for less than US $1,000 apiece. And the revolutions in biotech that have made such a feat possible are still gathering steam. Indeed, Earth BioGenome officials freely admit that their bold goal—to sequence those 1.8 million named species by 2035—won’t be possible without a hundredfold decrease in the time and cost of sequencing.
But the project’s success may ultimately hinge on functions other than sequencing. For example, after a creature’s genome is sequenced, the huge mass of raw genetic data—consisting of millions or billions of genetic building blocks called base pairs—must be annotated. That is, the tens of thousands of genes that make up the genome must be identified, located on chromosomes, and their functions or purpose described. And, of course, before an organism’s genome can be sequenced, its tissues must be sampled. To do that, researchers must locate the organism and, if it’s an animal, capture it. As I discovered with the Psyche team in the woods, valleys, and jagged peaks of South Tyrol, wrangling insects presents challenges that can defy logistics, technology, and even reason.
How Can You Explain the Surpassingly Strange Atlas Blue Butterfly?
When I first heard about Project Psyche, the first thing I wondered was, Why Lepidoptera I put the question to Charlotte Wright and Joana Meier at the hotel in Malles Venosta, Italy, that served as the headquarters for the Project Psyche expedition. They lead the project from its base at the Wellcome Sanger Institute in Cambridgeshire, England. The reasons, they tell me, span a range from pure science to completely commercial.

 At the Hotel Tyrol in the Italian Alps, lepidopterist Charlotte Wright of the Wellcome Sanger Institute, a leader of Project Psyche, dissects the yellow tiger moth captured near the stone hut. Packed with liquid nitrogen, the tissue samples will subsequently be sent to the institute in England for genome sequencing.Luigi Avantaggiato
At the Hotel Tyrol in the Italian Alps, lepidopterist Charlotte Wright of the Wellcome Sanger Institute, a leader of Project Psyche, dissects the yellow tiger moth captured near the stone hut. Packed with liquid nitrogen, the tissue samples will subsequently be sent to the institute in England for genome sequencing.Luigi Avantaggiato
The earliest Lepidoptera appeared 250 million to 300 million years ago. By studying and comparing the genomes of different species, Wright explains, “we can find out how they have evolved and how they’ve diversified, as there have been different climatic shifts in Europe. And the genomes can help to tell us why it is that some groups of Lepidoptera have evolved into a greater number of species than others.”
Those genomes will also offer insights into some of the most intriguing questions of evolutionary biology. Consider: Most moths and butterflies have genomes with around 31 pairs of chromosomes, which are the threadlike strands in every cell’s nucleus, each of which is a molecule of DNA. Collectively, chromosomes make up a creature’s genome. But a tiny minority of the Lepidoptera order have enormous numbers of chromosomes. Exhibit A is the Atlas blue butterfly, which has an astonishing 229 pairs of chromosomes.
The Atlas blue is “a very good example of something that’s really fascinating, but we cannot understand it just by looking at one species,” says Meier. “What we really need is what Psyche will provide, which is replications”—thousands of Lepidoptera genomes. And, not incidentally, the ability to browse them easily. “Then we will find many lineages that have an unusually large number of chromosomes, and we can then start to ask, ‘What changes each time? What do they have in common? Do they have a repair gene that’s broken?’ ”
 Some exceptional samples of Lepidoptera are preserved for entomological archives.Luigi Avantaggiato
Some exceptional samples of Lepidoptera are preserved for entomological archives.Luigi Avantaggiato
And it’s not just theoreticians eagerly awaiting such genomic data. One practical aspect of these studies has to do with moths’ impact on agriculture. “There’s billions and billions of euros lost because agriculturally, some species do a lot of damage,” says Meier.
Adds Wright, “Pests are moving to new regions where previously they weren’t present and causing huge losses because the crops there haven’t been developed to be protected against these new species.” The reasons why some species succeed in a new area as climate changes, and are able to adapt and thrive, are also understandable only by studying many genomes—of the creatures that succeed, as well as the ones that don’t. “It’s kind of a dynamic situation, of monitoring these pests’ movements,” says Wright.
 Shortly before sunset, Gioele Moro, of the Czech Academy of Sciences, sets up a moth trap on a mountain slope above the stone hut (the Alte Pforzheimer Hütte) in the Italian Alps. Luigi Avantaggiato
Shortly before sunset, Gioele Moro, of the Czech Academy of Sciences, sets up a moth trap on a mountain slope above the stone hut (the Alte Pforzheimer Hütte) in the Italian Alps. Luigi Avantaggiato
That, it turns out, takes a small army of grad students, researchers, and even citizen-scientists. Indeed, one of the goals of this expedition is to develop and refine best practices in collecting samples for genome sequencing and to train a cadre of young lepidopterists, who have varying levels of familiarity with the technologies of genome sequencing and annotation. On such techniques rests the success of not only Project Psyche, but also, ultimately, the Earth BioGenome Project.
To Catch a Moth, You’ve Got to Think Like One
It’s late in the afternoon of our first day in the high-altitude hut. Moro, of the Czech Academy of Sciences, is standing on a steeply raked mountainside in a dazzling sea of wildflowers—purple, yellow, lavender, crimson—that are gently swaying in the fading amber light. He’s wearing a black camp shirt, black cargo shorts, black socks, black hiking boots, and chunky retro eyewear, and he’s carrying a butterfly net (yep, it’s black). He’s still and silent, taking in nuances of light, vegetation, and wind that would affect a moth’s flight path through the area. Thinking like a moth, he visualizes the routes it would likely take through side valleys and ravines.
The objective is to figure out where to place three butterfly traps for the night. Setting the traps in different “microenvironments,” he explains, will likely yield a broader range of creatures. But there’s no formula for this. Capturing critters depends heavily on intuition arising from experience, perception, and judgment.
 Genetics researcher Noé Dogbo, of the Institute of Research on Insect Biology in Tours, France, chases a butterfly during a hunting session in the Roja mountains near Curon Venosta, Bolzano, Italy. Luigi Avantaggiato
Genetics researcher Noé Dogbo, of the Institute of Research on Insect Biology in Tours, France, chases a butterfly during a hunting session in the Roja mountains near Curon Venosta, Bolzano, Italy. Luigi Avantaggiato
“Over there”—he points across the valley to the opposite slope. “It faces north. See? No flowers. That’s what I mean by different microenvironments.” We’re perched on the south-facing slope, about 80 meters above the valley bottom, on a trail about as wide as a toaster oven.
Hours later, after dodging cow patties the size of dinner plates and gaping holes leading to marmot burrows, the locations are chosen and the traps are set. There’s one on the south slope, one on the north, and one near the fast-flowing stream between them. As the sky darkens to a deep blue, we trudge back to the hut to stoke the fire and wait.
At the crack of dawn the next day, Moro is jubilant as he returns with the night’s haul. There are at least 150 moths, including the spectacular yellow tiger moth. The species that are needed for Project Psyche, as identified by Torrado-Blanco, are put in plastic specimen jars and will make their way down to the makeshift lab at the Hotel Tyrol. There, they’ll be photographed and then stunned and killed by exposure to dry ice, before being dissected. The head, thorax, and abdomen will be packed in separate plastic tubes for state-of-the-art DNA and RNA sequencing at the laboratories of the Wellcome Sanger Institute. The Wellcome Trust is the lead sponsor of both Project Psyche and the Darwin Tree of Life project.

 Lepidopterist Joana Meier of the Wellcome Sanger Institute, a leader of Project Psyche, packs the abdomen of a moth into a vial for shipment from Italy to the institute in England. A bar code on the vial contains information about the sample and allows it to be tracked on its journey to the lab. Luigi Avantaggiato
Lepidopterist Joana Meier of the Wellcome Sanger Institute, a leader of Project Psyche, packs the abdomen of a moth into a vial for shipment from Italy to the institute in England. A bar code on the vial contains information about the sample and allows it to be tracked on its journey to the lab. Luigi Avantaggiato
The plastic tubes are packed in liquid-nitrogen-cooled shipping containers for the trip to Wellcome Sanger. DNA begins to break down almost immediately after death, especially in soft tissues. So the cryogenics are necessary to ensure that the samples arrive at Wellcome Sanger with as little degradation as possible.
Micromoths Are a Looming Challenge
Niklas Wahlberg of Lund University, in Sweden, is officially a “sampling hub leader” of Project Psyche. Unofficially, he’s one of the select few grizzled veterans here in Malles Venosta helping to mentor the young researchers, whose attendance is being funded through a European Union program called European Cooperation in Science and Technology.
 Niklas Wahlberg, an evolutionary biologist at Lund University in Sweden, captures a moth in a plastic container at a trapping site along an Alpine trail above Malles Venosta, Italy.Luigi Avantaggiato
Niklas Wahlberg, an evolutionary biologist at Lund University in Sweden, captures a moth in a plastic container at a trapping site along an Alpine trail above Malles Venosta, Italy.Luigi Avantaggiato
Wahlberg is an unabashed fan of moths. It’s not that he dislikes butterflies, mind you, it’s just that he’s a bit weary of them overshadowing moths in the public imagination. Butterflies are big, bright, and colorful, sure, but also delicate. They appeared much, much later than moths in evolutionary history. And they can’t even fly at night or in the rain. “Butterflies are just day-flying moths,” Wahlberg quips. “People think of them as different and special, but they’re not.”
In this new era of mass genome sequencing, they’re also arguably less important scientifically. To begin with, butterflies are just 10 percent of all known species of Lepidoptera—about 19,000 are butterflies while perhaps 180,000 or more are moths. Of the 11,000 European Lepidoptera species that are of interest to Project Psyche, only 560 of them are butterflies, by Wahlberg’s reckoning. And they’ve already collected two-thirds of them, he adds.
So the real challenge for Psyche is finding and identifying all those moths. Particularly the micromoths.
Micromoths have long vexed entomologists. The largest of them have wingspans about as wide as a U.S. dime, or a 2 euro cent coin; the smallest can fit on the head of a pin. As a group, they evolved not only much earlier than butterflies but also much earlier than all other moths (which are known as “macromoths”). There are a lot of micromoths—at least 62,000 species, by the current estimate. Among them are many pairs or other small groups of species that are so similar that not even the most experienced lepidopterists can tell them apart by eye.
 Charlotte Wright of the Wellcome Sanger Institute collects a moth at a light trap on an Alpine trail above Malles Venosta, Italy.Luigi Avantaggiato
Charlotte Wright of the Wellcome Sanger Institute collects a moth at a light trap on an Alpine trail above Malles Venosta, Italy.Luigi Avantaggiato
That’s going to be an enormous challenge for Project Psyche, Wahlberg notes. Fortunately, though, it’s a problem for which there is a technological solution: DNA barcoding.
Besides the DNA in the nuclei of every cell, there exists other genetic material, called mitochondrial DNA, outside of the nucleus. It’s relatively easy to access, and, crucially, there’s a mitochondrial gene, called CO1, that tends to vary markedly among species, even closely related ones. That makes this bit of genetic material invaluable for discriminating among related species. Researchers have built up several databases of these DNA barcodes that collectively contain millions of characteristic DNA sequences. “We have DNA barcodes for 99 percent of the Lepidoptera in Europe,” Wahlberg says. “And only about 5 percent of micromoth species have the same CO1 gene.”
DNA barcoding was invented in the early 2000s by Paul Hebert and colleagues at the University of Guelph, in Canada, and it has advanced greatly in recent years along with the DNA-sequencing technologies that underpin it. The technique starts with a minuscule sample of tissue; for example, in the makeshift lab at the hotel in Malles Venosta, researchers dissecting moths for sequencing also removed, for DNA barcoding, a leg of each moth whose species was not conclusively known.

 Staff Scientist Silvia Pérez Lluch of the Centre for Genomic Regulation in Barcelona retrieves tissue samples for genome sequencing. To minimize degradation of the DNA in the samples, they are stored at -80 °C.Luigi Avantaggiato
Staff Scientist Silvia Pérez Lluch of the Centre for Genomic Regulation in Barcelona retrieves tissue samples for genome sequencing. To minimize degradation of the DNA in the samples, they are stored at -80 °C.Luigi Avantaggiato
Genetic material is isolated from that tissue, and then a CO1 gene is “amplified,” or replicated into many millions of copies, using a standard biotechnical technique called polymerase chain reaction. That material is sequenced using any one of the dozen or more types of sequencing machines available to researchers.
For barcoding purposes, typical DNA sequences of the CO1 gene run between 400 and 800 base pairs. But lately researchers have been developing techniques that use shorter or longer barcodes. The shorter codes, called mini-barcodes, have proven more effective in identifying a species even when the DNA samples are incomplete or damaged. A mini-barcode might have 100 to 250 base pairs. Conversely, “super-barcodes,” which can be many thousands of base pairs, are useful for differentiating among closely related species—exactly the challenge with many of the micromoths.
Why RNA Will Make Annotating Faster
While the Psyche researchers honed the logistics and mechanics of sampling Lepidoptera, a different European Lepidoptera project was quietly making a technical advance that could resonate throughout the Earth BioGenome Project. Working together, Spanish and Andorran researchers affiliated with the Catalan Initiative for the Earth BioGenome Project sequenced the genome of the violet copper butterfly, Lycaena helle, a creature that was first studied in 1775. They described their efforts in a paper published by F1000Research.
This was no routine procedure. Typically when researchers map a genome, an organism is sampled and the DNA is sequenced. After sequencing, the mass of fragmented genetic data must be assembled into a complete genome sequence and then that complete sequence must be manually verified, in a process called curation, and then annotated. In annotation, the genome’s many genes are identified and, ideally, their functions described.
 Ivo Gut, director of Centro Nacional de Análisis Genómico in Barcelona, has high hopes for an emerging technique to identify the genes within a large mass of genetic data.Luigi Avantaggiato
Ivo Gut, director of Centro Nacional de Análisis Genómico in Barcelona, has high hopes for an emerging technique to identify the genes within a large mass of genetic data.Luigi Avantaggiato
Today, curation and annotation are time-consuming processes, regarded as major bottlenecks to the rapid progress that the Earth BioGenome Project desperately needs to reach its 2035 goal. Finding the thousands of genes within the huge mass of sequenced data is a mostly automated process now but it can involve some serious bioinformatic sleuthing. “You take your linear genome, your sequence, and you go and you say, ‘Ah, look here. There’s a gene that starts here,” says Ivo Gut, director of the Centro Nacional de Análisis Genómico (CNAG), in Barcelona. “ ‘And this is the structure of the gene.’ And then you can sort of figure out what that is. You look whether that gene is known, for example, in another species. And then you go to the next one, and so on. And just by these similarity searches, you can usually annotate almost 80 percent, or maybe 70 percent,” of what are known as coding genes in the genome. These coding genes encode the many proteins produced by cells, which serve vital functions in the organism.
Gut also notes that to perform annotations researchers are making increasing use of another genetic molecule, RNA, or ribonucleic acid. When a gene creates, or “expresses,” a protein, RNA acts as the “messenger,” carrying the genetic code outside of the cell nucleus to the protein-making apparatus of the cell. Therefore RNA is extremely useful in figuring out where the protein-coding genes are in the genome. Different cells in the body express different proteins, but in every case that expression occurs because of a specific gene, and that gene can be identified conclusively from the RNA associated with it.
The breakthrough in the research by the Spanish and Andorran researchers was using a technique called long-read sequencing to sequence all of the RNA in their samples. While sequencing a genome, long-read machines handle much longer segments of DNA than traditional short-read systems. The greater length confers several advantages, including the ability to easily resolve repetitive sequences that can trip up short-read machines. [For more on long-read genome sequencing, see my recent article “The Quest to Sequence the Genomes of Everything, in IEEE Spectrum.”] The researchers came from four Barcelona organizations—CNAG, the Centre for Genomic Regulation (CRG), the Institute of Evolutionary Biology at Pompeu Fabra University, and the University of Barcelona—and from Andorra Research and Innovation, in Sant Julià de Lòria.
The genome of the female violet copper butterfly, which inhabits a huge swath of territory stretching from the Pyrenees to Siberia, consists of 25 pairs of chromosomes with a total of 547,306,268 base pairs. By using long-read sequencing of the RNA in the sample, the researchers were able to identify 20,122 protein-coding genes and 4,264 noncoding genes. In contrast to protein-coding genes, noncoding genes are harder to identify from one species to the next and they are also very difficult to predict by computational means. Many noncoding genes serve important regulatory, protective, or other functions within a cell. Yet at least 30 percent of all annotated Lepidopteran genomes produced so far lack annotations of noncoding genes, and those that include them generally count relatively few, says Roderic Guigó Serra, who leads the Bioinformatics and Genomics program at the CRG.
“Long-read RNA sequencing may be the only way to precisely locate them in genome sequences,” he says. With long-read RNA sequencing, “we get better information on where the genes are and a more precise definition of the boundaries of the genes, and also we see genes that had not been seen before,” Serra declares.
 At the Guigò Lab of the Centre for Genomic Regulation in Barcelona, a technician loads a sample into a genome sequencing machine. Luigi Avantaggiato
At the Guigò Lab of the Centre for Genomic Regulation in Barcelona, a technician loads a sample into a genome sequencing machine. Luigi Avantaggiato
His group is now applying the long-read RNA sequencing technique to a host of other species—including humans. They’re doing this through Gencode, an international consortium that aims to produce improved, “reference” annotations for the human and mouse genomes. Twenty-five years after the first draft sequence of the human genome, it turns out that there are still gaps in it—particularly regarding the noncoding genes. Recently, using long-read RNA sequencing, the Gencode team shocked biologists by identifying 18,000 previously unknown noncoding human genes. “These genes have been essentially ignored for almost 25 years, underscoring the power of the long-read RNA sequencing technology,” says Serra.
Researchers are counting on such advances to help power them in their grand quest of sequencing and annotating the world’s organisms. And within that quest, Project Psyche is off to an encouraging start. With nearly 3,000 of Europe’s 11,000 Lepidopteran species sampled and more than 1,000 of those sequenced, Lepidoptera are now the most widely sequenced order of organisms. Still, that leaves perhaps 170,000 other members of the order elsewhere in the world to be sampled and sequenced.
It’s a mammoth task. As they grapple with it, its practitioners can take inspiration from the novelist and lepidopterist Vladimir Nabokov. “My loathings are simple,” he wrote in 1973. “Stupidity, oppression, crime, cruelty, soft music. My pleasures are the most intense known to man: writing and butterfly hunting.”




 The Alte Pforzheimer Hütte, a stone house originally built in 1901, served as a base camp for the lepidopterists hunting rare moths in the Italian Alps.Luigi Avantaggiato
The Alte Pforzheimer Hütte, a stone house originally built in 1901, served as a base camp for the lepidopterists hunting rare moths in the Italian Alps.Luigi Avantaggiato A few of the scores of moths captured on a single night at a site in the Italian Alps are lined up on a bench in the stone hut. Researchers will identify the moths’ species and some of the insects will be sent on for tissue sampling and eventual genome sequencing. Luigi Avantaggiato
A few of the scores of moths captured on a single night at a site in the Italian Alps are lined up on a bench in the stone hut. Researchers will identify the moths’ species and some of the insects will be sent on for tissue sampling and eventual genome sequencing. Luigi Avantaggiato A yellow tiger moth, Arctia flavia, is among the catch at the stone hut, at an altitude of 2,300 meters.
A yellow tiger moth, Arctia flavia, is among the catch at the stone hut, at an altitude of 2,300 meters.  At the crack of dawn in the stone hut, researchers [from left] Eric Toro Delgado, Laura Torrado-Blanco, Mónica Doblas-Bajo, and Gioele Moro (standing) unpack and examine the moths captured during the previous night.Luigi Avantaggiato
At the crack of dawn in the stone hut, researchers [from left] Eric Toro Delgado, Laura Torrado-Blanco, Mónica Doblas-Bajo, and Gioele Moro (standing) unpack and examine the moths captured during the previous night.Luigi Avantaggiato
 At the Hotel Tyrol in the Italian Alps, lepidopterist Charlotte Wright of the Wellcome Sanger Institute, a leader of Project Psyche, dissects the yellow tiger moth captured near the stone hut. Packed with liquid nitrogen, the tissue samples will subsequently be sent to the institute in England for genome sequencing.Luigi Avantaggiato
At the Hotel Tyrol in the Italian Alps, lepidopterist Charlotte Wright of the Wellcome Sanger Institute, a leader of Project Psyche, dissects the yellow tiger moth captured near the stone hut. Packed with liquid nitrogen, the tissue samples will subsequently be sent to the institute in England for genome sequencing.Luigi Avantaggiato Some exceptional samples of Lepidoptera are preserved for entomological archives.Luigi Avantaggiato
Some exceptional samples of Lepidoptera are preserved for entomological archives.Luigi Avantaggiato Shortly before sunset, Gioele Moro, of the Czech Academy of Sciences, sets up a moth trap on a mountain slope above the stone hut (the Alte Pforzheimer Hütte) in the Italian Alps. Luigi Avantaggiato
Shortly before sunset, Gioele Moro, of the Czech Academy of Sciences, sets up a moth trap on a mountain slope above the stone hut (the Alte Pforzheimer Hütte) in the Italian Alps. Luigi Avantaggiato  Genetics researcher Noé Dogbo, of the Institute of Research on Insect Biology in Tours, France, chases a butterfly during a hunting session in the Roja mountains near Curon Venosta, Bolzano, Italy. Luigi Avantaggiato
Genetics researcher Noé Dogbo, of the Institute of Research on Insect Biology in Tours, France, chases a butterfly during a hunting session in the Roja mountains near Curon Venosta, Bolzano, Italy. Luigi Avantaggiato 
 Lepidopterist Joana Meier of the Wellcome Sanger Institute, a leader of Project Psyche, packs the abdomen of a moth into a vial for shipment from Italy to the institute in England. A bar code on the vial contains information about the sample and allows it to be tracked on its journey to the lab. Luigi Avantaggiato
Lepidopterist Joana Meier of the Wellcome Sanger Institute, a leader of Project Psyche, packs the abdomen of a moth into a vial for shipment from Italy to the institute in England. A bar code on the vial contains information about the sample and allows it to be tracked on its journey to the lab. Luigi Avantaggiato Niklas Wahlberg, an evolutionary biologist at Lund University in Sweden, captures a moth in a plastic container at a trapping site along an Alpine trail above Malles Venosta, Italy.Luigi Avantaggiato
Niklas Wahlberg, an evolutionary biologist at Lund University in Sweden, captures a moth in a plastic container at a trapping site along an Alpine trail above Malles Venosta, Italy.Luigi Avantaggiato Charlotte Wright of the Wellcome Sanger Institute collects a moth at a light trap on an Alpine trail above Malles Venosta, Italy.Luigi Avantaggiato
Charlotte Wright of the Wellcome Sanger Institute collects a moth at a light trap on an Alpine trail above Malles Venosta, Italy.Luigi Avantaggiato
 Staff Scientist Silvia Pérez Lluch of the Centre for Genomic Regulation in Barcelona retrieves tissue samples for genome sequencing. To minimize degradation of the DNA in the samples, they are stored at -80 °C.Luigi Avantaggiato
Staff Scientist Silvia Pérez Lluch of the Centre for Genomic Regulation in Barcelona retrieves tissue samples for genome sequencing. To minimize degradation of the DNA in the samples, they are stored at -80 °C.Luigi Avantaggiato Ivo Gut, director of Centro Nacional de Análisis Genómico in Barcelona, has high hopes for an emerging technique to identify the genes within a large mass of genetic data.Luigi Avantaggiato
Ivo Gut, director of Centro Nacional de Análisis Genómico in Barcelona, has high hopes for an emerging technique to identify the genes within a large mass of genetic data.Luigi Avantaggiato At the Guigò Lab of the Centre for Genomic Regulation in Barcelona, a technician loads a sample into a genome sequencing machine. Luigi Avantaggiato
At the Guigò Lab of the Centre for Genomic Regulation in Barcelona, a technician loads a sample into a genome sequencing machine. Luigi Avantaggiato















 These series of images show how the robot moves through the stomach. In the rightmost column, the researchers also demonstrate targeted drug delivery.
These series of images show how the robot moves through the stomach. In the rightmost column, the researchers also demonstrate targeted drug delivery.